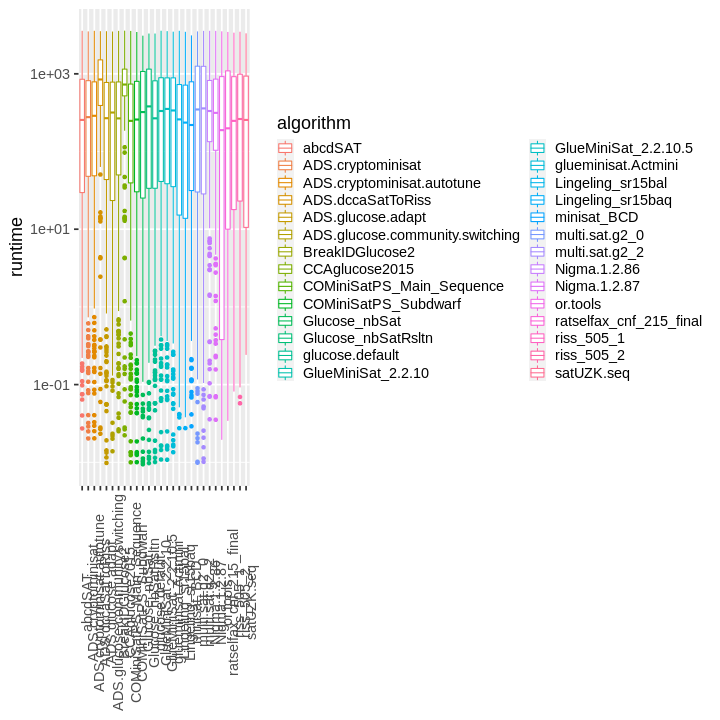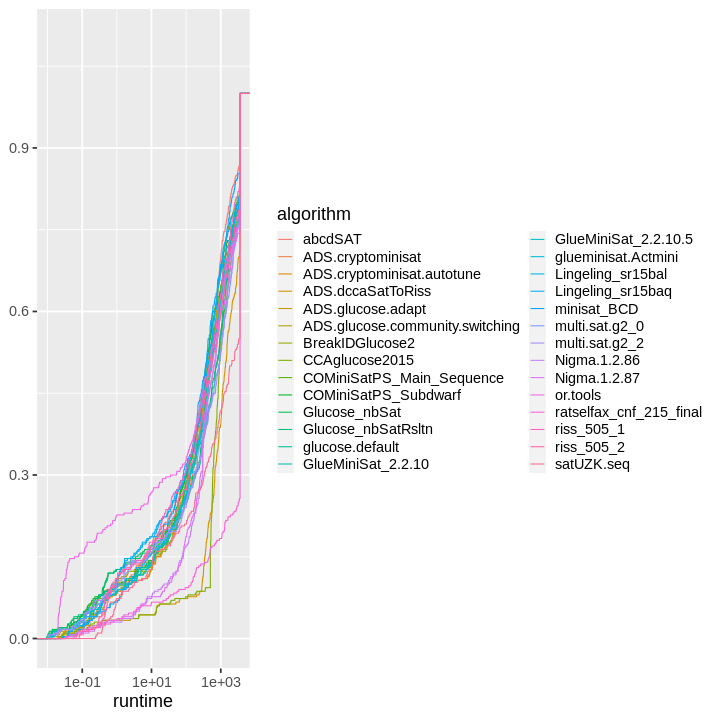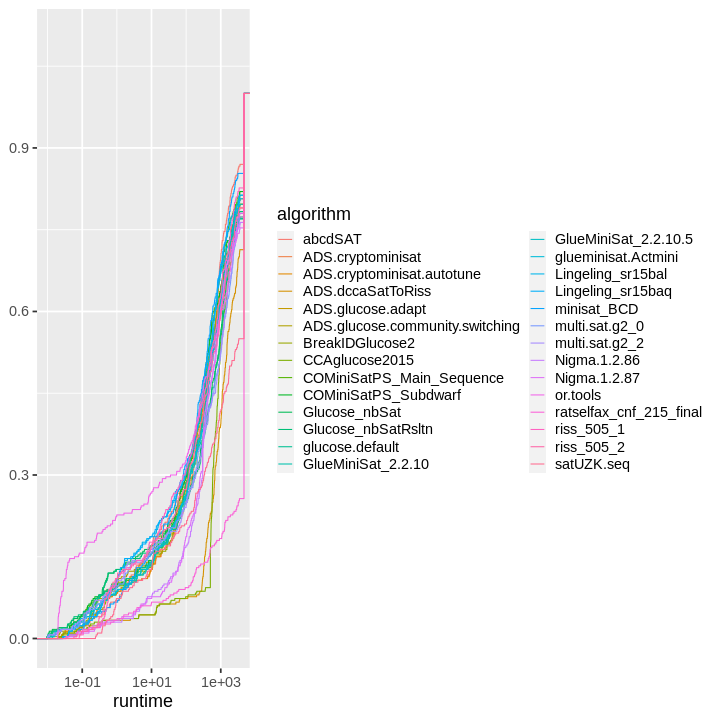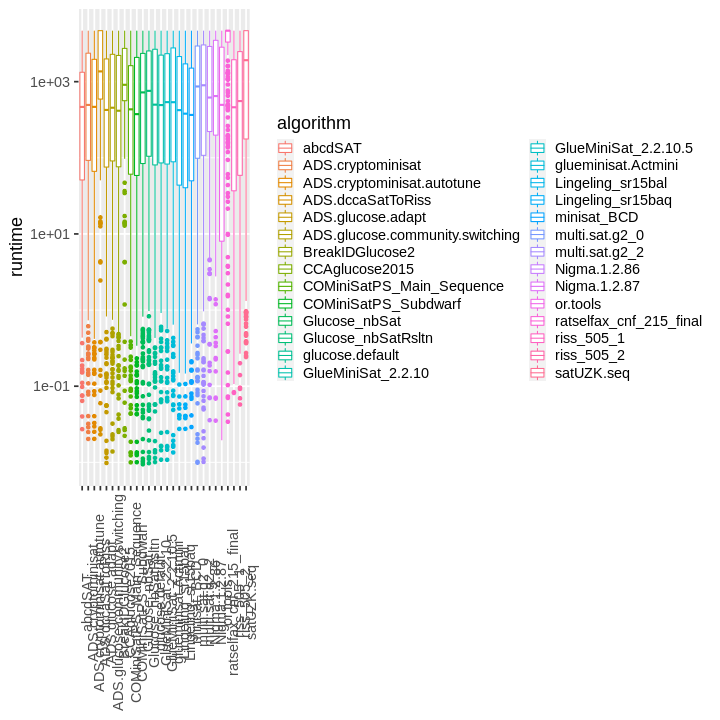
| obs | nas | min | qu_1st | med | mean | qu_3rd | max | sd | coeff_var | |
|---|---|---|---|---|---|---|---|---|---|---|
| ADS.cryptominisat | 300 | 0 | 0.020286 | 92.417 | 494.9 | 1216.92 | 2367.33 | 3600 | 1397.35 | 1.14826 |
| ADS.cryptominisat.autotune | 300 | 0 | 0.0203381 | 66.4585 | 466.548 | 1169.65 | 1964.98 | 3600 | 1372.11 | 1.1731 |
| ADS.dccaSatToRiss | 300 | 0 | 0.0233011 | 591.004 | 1364.4 | 1809.34 | 3600 | 3600 | 1369.38 | 0.756839 |
| ADS.glucose.adapt | 300 | 0 | 0.009817 | 75.6422 | 425.591 | 1174.2 | 1988.92 | 3600 | 1387.9 | 1.182 |
| ADS.glucose.community.switching | 300 | 0 | 0.013876 | 90.9123 | 454.408 | 1256.39 | 2284.29 | 3600 | 1422 | 1.13182 |
| BreakIDGlucose2 | 300 | 0 | 0.0251101 | 76.1845 | 415.057 | 1190.56 | 2209.03 | 3600 | 1400.3 | 1.17617 |
| CCAglucose2015 | 300 | 0 | 0.022267 | 562.385 | 909.917 | 1540.55 | 2734.03 | 3600 | 1279.95 | 0.830838 |
| COMiniSatPS_Main_Sequence | 300 | 0 | 0.00999496 | 60.9539 | 432.935 | 1084.71 | 1621.88 | 3600 | 1351.89 | 1.24632 |
| COMiniSatPS_Subdwarf | 300 | 0 | 0.0100371 | 58.5645 | 374.43 | 1131.36 | 2081.94 | 3600 | 1377.92 | 1.21793 |
| Glucose_nbSat | 300 | 0 | 0.00942791 | 84.06 | 722.605 | 1307.27 | 2350.39 | 3600 | 1398.36 | 1.06968 |
| Glucose_nbSatRsltn | 300 | 0 | 0.00975408 | 104.74 | 757.635 | 1326.78 | 2535.22 | 3600 | 1385.81 | 1.04449 |
| GlueMiniSat_2.2.10 | 300 | 0 | 0.010843 | 85.629 | 493.322 | 1186.32 | 2232.21 | 3600 | 1366.35 | 1.15176 |
| GlueMiniSat_2.2.10.5 | 300 | 0 | 0.0107971 | 82.7613 | 538.103 | 1224.51 | 2333.24 | 3600 | 1384 | 1.13025 |
| Lingeling_sr15bal | 300 | 0 | 0.0274709 | 43.8066 | 422.921 | 1146.65 | 2139.04 | 3600 | 1398.76 | 1.21987 |
| Lingeling_sr15baq | 300 | 0 | 0.0275191 | 40.4331 | 380.214 | 1089.31 | 1715.44 | 3600 | 1371.67 | 1.25921 |
| Nigma.1.2.86 | 300 | 0 | 0.0357599 | 221.212 | 619.452 | 1327.78 | 2898.77 | 3600 | 1416.93 | 1.06714 |
| Nigma.1.2.87 | 300 | 0 | 0.0352739 | 198.592 | 646.966 | 1349.05 | 3493.95 | 3600 | 1436.38 | 1.06473 |
| abcdSAT | 300 | 0 | 0.0272769 | 51.178 | 466.839 | 977.358 | 1328.7 | 3600 | 1225.85 | 1.25425 |
| glucose.default | 300 | 0 | 0.0100621 | 80.6243 | 498.45 | 1260.08 | 2667.05 | 3600 | 1436.31 | 1.13986 |
| glueminisat.Actmini | 300 | 0 | 0.0134831 | 88.5993 | 537.454 | 1280.5 | 2775.87 | 3600 | 1427.2 | 1.11456 |
| minisat_BCD | 300 | 0 | 0.0290739 | 49.8396 | 365.713 | 996.284 | 1507.14 | 3600 | 1270.11 | 1.27485 |
| multi.sat.g2_0 | 300 | 0 | 0.00986789 | 98.6871 | 864.628 | 1417.8 | 2939.51 | 3600 | 1430.99 | 1.0093 |
| multi.sat.g2_2 | 300 | 0 | 0.0101351 | 107.833 | 897.061 | 1424.59 | 3040.63 | 3600 | 1431.36 | 1.00475 |
| or.tools | 300 | 0 | 0.0194899 | 8.03079 | 495.793 | 1283.99 | 2836.37 | 3600 | 1458.51 | 1.13592 |
| ratselfax_cnf_215_final | 300 | 0 | 0.0341411 | 3310.59 | 3600 | 2856.58 | 3600 | 3600 | 1355.9 | 0.474659 |
| riss_505_1 | 300 | 0 | 0.0814799 | 36.8773 | 461.437 | 1133.31 | 1941.55 | 3600 | 1342.93 | 1.18496 |
| riss_505_2 | 300 | 0 | 0.057448 | 58.8041 | 556.543 | 1262.05 | 2484.22 | 3600 | 1410.81 | 1.11788 |
| satUZK.seq | 300 | 0 | 0.241177 | 176.75 | 1916.25 | 1968.55 | 3600 | 3600 | 1601.48 | 0.813534 |
| ok | timeout | memout | not_applicable | crash | other | |
|---|---|---|---|---|---|---|
| abcdSAT | 87.000 | 13.000 | 0.000 | 0.000 | 0.000 | 0.000 |
| ADS.cryptominisat | 79.000 | 21.000 | 0.000 | 0.000 | 0.000 | 0.000 |
| ADS.cryptominisat.autotune | 80.667 | 19.333 | 0.000 | 0.000 | 0.000 | 0.000 |
| ADS.dccaSatToRiss | 71.333 | 28.667 | 0.000 | 0.000 | 0.000 | 0.000 |
| ADS.glucose.adapt | 79.667 | 20.333 | 0.000 | 0.000 | 0.000 | 0.000 |
| ADS.glucose.community.switching | 78.333 | 21.667 | 0.000 | 0.000 | 0.000 | 0.000 |
| BreakIDGlucose2 | 79.667 | 20.333 | 0.000 | 0.000 | 0.000 | 0.000 |
| CCAglucose2015 | 77.333 | 22.667 | 0.000 | 0.000 | 0.000 | 0.000 |
| COMiniSatPS_Main_Sequence | 82.000 | 18.000 | 0.000 | 0.000 | 0.000 | 0.000 |
| COMiniSatPS_Subdwarf | 82.000 | 18.000 | 0.000 | 0.000 | 0.000 | 0.000 |
| Glucose_nbSat | 77.000 | 23.000 | 0.000 | 0.000 | 0.000 | 0.000 |
| Glucose_nbSatRsltn | 78.333 | 21.667 | 0.000 | 0.000 | 0.000 | 0.000 |
| glucose.default | 77.000 | 23.000 | 0.000 | 0.000 | 0.000 | 0.000 |
| GlueMiniSat_2.2.10 | 81.333 | 18.667 | 0.000 | 0.000 | 0.000 | 0.000 |
| GlueMiniSat_2.2.10.5 | 79.667 | 20.333 | 0.000 | 0.000 | 0.000 | 0.000 |
| glueminisat.Actmini | 78.000 | 22.000 | 0.000 | 0.000 | 0.000 | 0.000 |
| Lingeling_sr15bal | 80.667 | 19.333 | 0.000 | 0.000 | 0.000 | 0.000 |
| Lingeling_sr15baq | 81.333 | 18.667 | 0.000 | 0.000 | 0.000 | 0.000 |
| minisat_BCD | 85.333 | 14.667 | 0.000 | 0.000 | 0.000 | 0.000 |
| multi.sat.g2_0 | 77.333 | 22.667 | 0.000 | 0.000 | 0.000 | 0.000 |
| multi.sat.g2_2 | 76.333 | 23.667 | 0.000 | 0.000 | 0.000 | 0.000 |
| Nigma.1.2.86 | 76.333 | 23.667 | 0.000 | 0.000 | 0.000 | 0.000 |
| Nigma.1.2.87 | 75.333 | 24.667 | 0.000 | 0.000 | 0.000 | 0.000 |
| or.tools | 78.000 | 22.000 | 0.000 | 0.000 | 0.000 | 0.000 |
| ratselfax_cnf_215_final | 25.667 | 74.333 | 0.000 | 0.000 | 0.000 | 0.000 |
| riss_505_1 | 82.667 | 17.333 | 0.000 | 0.000 | 0.000 | 0.000 |
| riss_505_2 | 79.000 | 21.000 | 0.000 | 0.000 | 0.000 | 0.000 |
| satUZK.seq | 55.000 | 45.000 | 0.000 | 0.000 | 0.000 | 0.000 |

## Warning: Removed 1976 rows containing non-finite values (stat_boxplot).