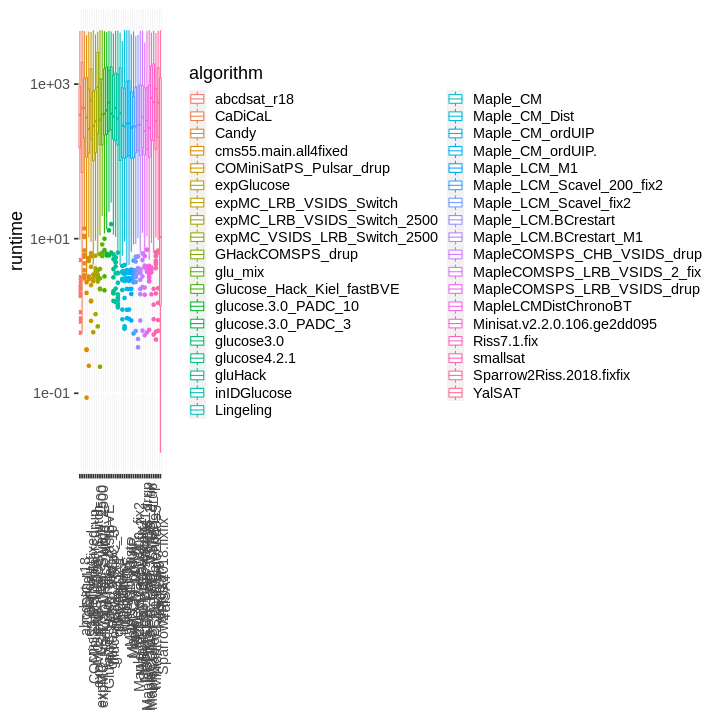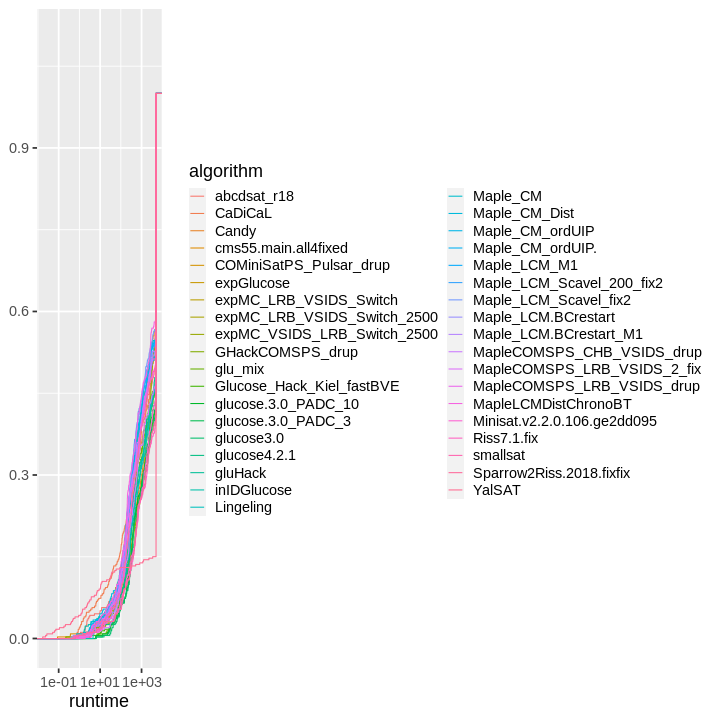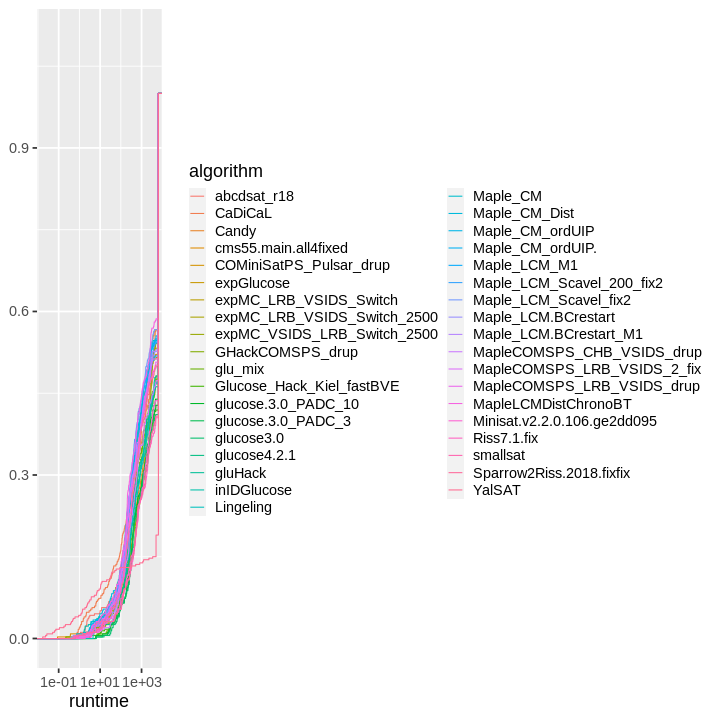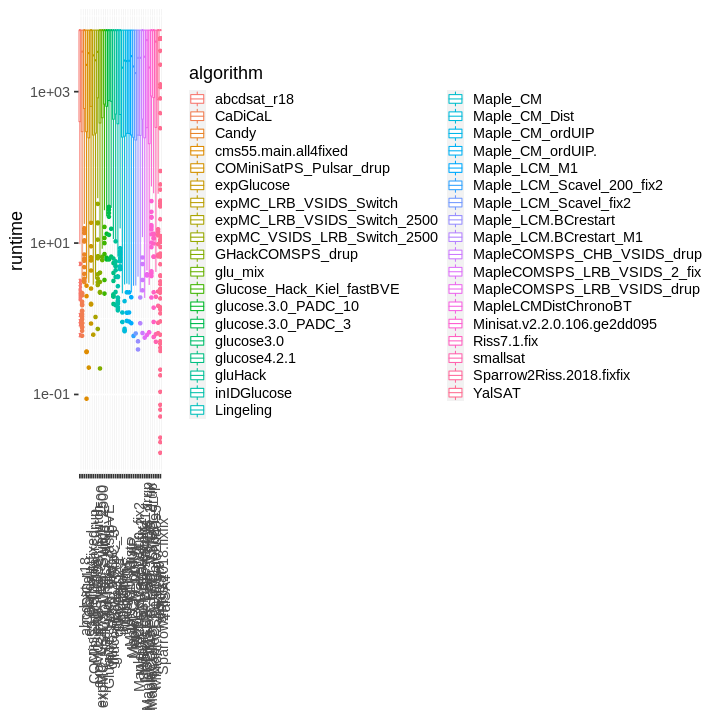
| obs | nas | min | qu_1st | med | mean | qu_3rd | max | sd | coeff_var | |
|---|---|---|---|---|---|---|---|---|---|---|
| abcdsat_r18 | 353 | 0 | 0.607502 | 401.026 | 5000.04 | 2905.42 | 5001.01 | 5001.02 | 2225.27 | 0.765903 |
| CaDiCaL | 353 | 0 | 0.595971 | 295.856 | 3379.47 | 2813.53 | 5001.01 | 5001.01 | 2183.59 | 0.776101 |
| Candy | 353 | 0 | 3.05981 | 598.512 | 5000.3 | 3273.41 | 5001.01 | 5001.01 | 2142.7 | 0.654577 |
| cms55.main.all4fixed | 353 | 0 | 0.0878539 | 295.716 | 2257.04 | 2647.31 | 5000.66 | 5001.01 | 2195.92 | 0.829489 |
| COMiniSatPS_Pulsar_drup | 353 | 0 | 0.22654 | 245.378 | 3224.43 | 2772.88 | 5000.66 | 5001.02 | 2251.83 | 0.81209 |
| expGlucose | 353 | 0 | 0.863128 | 638.242 | 5000.1 | 3160.99 | 5001.01 | 5001.01 | 2115.09 | 0.669122 |
| YalSAT | 353 | 0 | 0.016957 | 5000.06 | 5000.33 | 4278.8 | 5000.57 | 5001.01 | 1734.9 | 0.405464 |
| smallsat | 353 | 0 | 0.496339 | 337.374 | 4585.72 | 2858.81 | 5001.01 | 5001.02 | 2239.94 | 0.783522 |
| Maple_LCM_Scavel_200_fix2 | 353 | 0 | 0.593495 | 249.338 | 2152.4 | 2633.62 | 5000.62 | 5001.01 | 2276.29 | 0.864319 |
| Maple_LCM_Scavel_fix2 | 353 | 0 | 0.640118 | 229.801 | 1761.32 | 2553.79 | 5000.62 | 5001.01 | 2247.29 | 0.879984 |
| Maple_LCM_M1 | 353 | 0 | 0.874415 | 267.557 | 2947.35 | 2758.99 | 5000.63 | 5001.01 | 2260.34 | 0.819263 |
| Maple_LCM.BCrestart_M1 | 353 | 0 | 0.87234 | 263.926 | 2797.68 | 2738.17 | 5000.62 | 5001.02 | 2235.69 | 0.816492 |
| Maple_LCM.BCrestart | 353 | 0 | 0.393768 | 266.31 | 2794.76 | 2759.61 | 5000.69 | 5001.01 | 2234.17 | 0.809598 |
| Maple_CM_ordUIP. | 353 | 0 | 0.91849 | 280.165 | 2553.84 | 2678.01 | 5000.56 | 5001.02 | 2225.36 | 0.830975 |
| Maple_CM_ordUIP | 353 | 0 | 0.936933 | 281.154 | 2536.13 | 2676.37 | 5000.65 | 5001.01 | 2225.29 | 0.831458 |
| Maple_CM_Dist | 353 | 0 | 0.684711 | 258.113 | 2574.19 | 2654.3 | 5000.61 | 5001.01 | 2255.31 | 0.849681 |
| Maple_CM | 353 | 0 | 0.906671 | 250.732 | 2075.64 | 2617.72 | 5000.61 | 5001.01 | 2251.18 | 0.859976 |
| MapleLCMDistChronoBT | 353 | 0 | 0.662318 | 205.195 | 2080.17 | 2520.49 | 5000.57 | 5001.01 | 2230.91 | 0.88511 |
| Lingeling | 353 | 0 | 4.52935 | 498.267 | 5000.26 | 3112.54 | 5001.01 | 5001.02 | 2162.57 | 0.694794 |
| inIDGlucose | 353 | 0 | 1.29105 | 375.871 | 5000.11 | 3082.37 | 5001.01 | 5001.02 | 2189.18 | 0.710227 |
| glu_mix | 353 | 0 | 5.96379 | 452.413 | 5000.29 | 3080.3 | 5001.01 | 5001.01 | 2206.75 | 0.716409 |
| gluHack | 353 | 0 | 1.51409 | 504.895 | 5000.19 | 3064.04 | 5001.01 | 5001.01 | 2171.37 | 0.708664 |
| Glucose_Hack_Kiel_fastBVE | 353 | 0 | 2.1517 | 683.839 | 5000.67 | 3334.09 | 5001.01 | 5001.02 | 2098.77 | 0.629486 |
| glucose.3.0_PADC_3 | 353 | 0 | 6.07533 | 762.768 | 5000.34 | 3320.49 | 5001.01 | 5001.11 | 2093.24 | 0.630401 |
| glucose.3.0_PADC_10 | 353 | 0 | 6.07683 | 639.892 | 5000.39 | 3258.42 | 5001.01 | 5001.08 | 2109.88 | 0.647515 |
| glucose4.2.1 | 353 | 0 | 1.56823 | 431.539 | 5000.24 | 3121.44 | 5001.01 | 5001.01 | 2168.45 | 0.694696 |
| expMC_VSIDS_LRB_Switch_2500 | 353 | 0 | 0.733243 | 827.828 | 3355.09 | 3058.98 | 5000.55 | 5001.01 | 2029.96 | 0.663607 |
| expMC_LRB_VSIDS_Switch_2500 | 353 | 0 | 1.05537 | 282.667 | 2596.21 | 2714.94 | 5000.7 | 5001.01 | 2247.06 | 0.827664 |
| expMC_LRB_VSIDS_Switch | 353 | 0 | 0.616435 | 266.861 | 3013.88 | 2764.51 | 5000.62 | 5001.01 | 2257.93 | 0.816755 |
| GHackCOMSPS_drup | 353 | 0 | 0.22072 | 385.013 | 5000.14 | 3030.76 | 5000.74 | 5001.01 | 2203.11 | 0.726915 |
| glucose3.0 | 353 | 0 | 6.6208 | 744.573 | 5000.56 | 3367.83 | 5001.01 | 5001.01 | 2094.24 | 0.621839 |
| MapleCOMSPS_CHB_VSIDS_drup | 353 | 0 | 0.647167 | 433.445 | 5000.11 | 3022.82 | 5000.68 | 5001.01 | 2219.74 | 0.734326 |
| MapleCOMSPS_LRB_VSIDS_2_fix | 353 | 0 | 0.563273 | 245.528 | 2889.25 | 2759.01 | 5000.7 | 5001.02 | 2258.83 | 0.81871 |
| MapleCOMSPS_LRB_VSIDS_drup | 353 | 0 | 0.612203 | 334.58 | 4798.02 | 2850.02 | 5000.7 | 5001.01 | 2248.31 | 0.788876 |
| Minisat.v2.2.0.106.ge2dd095 | 353 | 0 | 1.9075 | 875.121 | 5000.53 | 3294.99 | 5001.01 | 5001.01 | 2079.09 | 0.630986 |
| Riss7.1.fix | 353 | 0 | 0.574338 | 834.757 | 5000.67 | 3439.65 | 5001.01 | 5001.02 | 2067.44 | 0.601063 |
| Sparrow2Riss.2018.fixfix | 353 | 0 | 0.645421 | 782.342 | 5000.68 | 3402.2 | 5001.01 | 5001.01 | 2076.1 | 0.610222 |
| ok | timeout | memout | not_applicable | crash | other | |
|---|---|---|---|---|---|---|
| abcdsat_r18 | 49.858 | 50.142 | 0.000 | 0.000 | 0.000 | 0.000 |
| CaDiCaL | 56.657 | 43.343 | 0.000 | 0.000 | 0.000 | 0.000 |
| Candy | 42.776 | 57.224 | 0.000 | 0.000 | 0.000 | 0.000 |
| cms55.main.all4fixed | 56.657 | 43.343 | 0.000 | 0.000 | 0.000 | 0.000 |
| COMiniSatPS_Pulsar_drup | 52.125 | 47.875 | 0.000 | 0.000 | 0.000 | 0.000 |
| expGlucose | 46.742 | 53.258 | 0.000 | 0.000 | 0.000 | 0.000 |
| expMC_LRB_VSIDS_Switch | 51.558 | 48.442 | 0.000 | 0.000 | 0.000 | 0.000 |
| expMC_LRB_VSIDS_Switch_2500 | 53.258 | 46.742 | 0.000 | 0.000 | 0.000 | 0.000 |
| expMC_VSIDS_LRB_Switch_2500 | 54.108 | 45.892 | 0.000 | 0.000 | 0.000 | 0.000 |
| GHackCOMSPS_drup | 48.159 | 51.841 | 0.000 | 0.000 | 0.000 | 0.000 |
| glu_mix | 47.592 | 52.408 | 0.000 | 0.000 | 0.000 | 0.000 |
| Glucose_Hack_Kiel_fastBVE | 41.926 | 58.074 | 0.000 | 0.000 | 0.000 | 0.000 |
| glucose.3.0_PADC_10 | 43.909 | 56.091 | 0.000 | 0.000 | 0.000 | 0.000 |
| glucose.3.0_PADC_3 | 42.776 | 57.224 | 0.000 | 0.000 | 0.000 | 0.000 |
| glucose3.0 | 41.076 | 58.924 | 0.000 | 0.000 | 0.000 | 0.000 |
| glucose4.2.1 | 46.176 | 53.824 | 0.000 | 0.000 | 0.000 | 0.000 |
| gluHack | 48.159 | 51.841 | 0.000 | 0.000 | 0.000 | 0.000 |
| inIDGlucose | 47.025 | 52.975 | 0.000 | 0.000 | 0.000 | 0.000 |
| Lingeling | 46.742 | 53.258 | 0.000 | 0.000 | 0.000 | 0.000 |
| Maple_CM | 54.674 | 45.326 | 0.000 | 0.000 | 0.000 | 0.000 |
| Maple_CM_Dist | 54.391 | 45.609 | 0.000 | 0.000 | 0.000 | 0.000 |
| Maple_CM_ordUIP | 55.524 | 44.476 | 0.000 | 0.000 | 0.000 | 0.000 |
| Maple_CM_ordUIP. | 54.958 | 45.042 | 0.000 | 0.000 | 0.000 | 0.000 |
| Maple_LCM_M1 | 51.841 | 48.159 | 0.000 | 0.000 | 0.000 | 0.000 |
| Maple_LCM_Scavel_200_fix2 | 54.391 | 45.609 | 0.000 | 0.000 | 0.000 | 0.000 |
| Maple_LCM_Scavel_fix2 | 56.657 | 43.343 | 0.000 | 0.000 | 0.000 | 0.000 |
| Maple_LCM.BCrestart | 52.691 | 47.309 | 0.000 | 0.000 | 0.000 | 0.000 |
| Maple_LCM.BCrestart_M1 | 52.975 | 47.025 | 0.000 | 0.000 | 0.000 | 0.000 |
| MapleCOMSPS_CHB_VSIDS_drup | 47.309 | 52.691 | 0.000 | 0.000 | 0.000 | 0.000 |
| MapleCOMSPS_LRB_VSIDS_2_fix | 51.275 | 48.725 | 0.000 | 0.000 | 0.000 | 0.000 |
| MapleCOMSPS_LRB_VSIDS_drup | 50.708 | 49.292 | 0.000 | 0.000 | 0.000 | 0.000 |
| MapleLCMDistChronoBT | 58.640 | 41.360 | 0.000 | 0.000 | 0.000 | 0.000 |
| Minisat.v2.2.0.106.ge2dd095 | 43.626 | 56.374 | 0.000 | 0.000 | 0.000 | 0.000 |
| Riss7.1.fix | 40.510 | 59.490 | 0.000 | 0.000 | 0.000 | 0.000 |
| smallsat | 50.142 | 49.858 | 0.000 | 0.000 | 0.000 | 0.000 |
| Sparrow2Riss.2018.fixfix | 40.793 | 59.207 | 0.000 | 0.000 | 0.000 | 0.000 |
| YalSAT | 18.980 | 81.020 | 0.000 | 0.000 | 0.000 | 0.000 |

## Warning: Removed 6681 rows containing non-finite values (stat_boxplot).